Molecular Modeling Helps Drug Researchers

Latest News
January 20, 2015
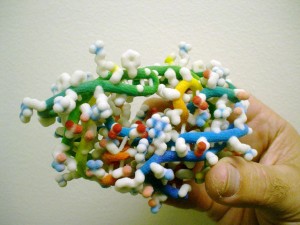 A model of the HIV protease, a target for AIDS therapy. Scientists at Scripps Molecular Graphics Laboratory use models to help design new drug candidates. Image: Scripps
A model of the HIV protease, a target for AIDS therapy. Scientists at Scripps Molecular Graphics Laboratory use models to help design new drug candidates. Image: ScrippsProtein folding is a complex process through which protein structures assume their functional shape. Studying this process is a critical part of molecular biology, and can help provide better insight into diseases and how to treat them, among other things. These types of molecules have typically been studied using 2D software renderings, but a number of facilities have turned to 3D printing to improve their understanding of molecular structures.
Arthur Olson, a molecular biologist at the Molecular Graphics Laboratory at the Scripps Research Institute in La Jolla, CA, for instance is using 3D printed molecule models and augmented reality technology to study the HIV virus, and is sharing those models with researchers via the National Institutes of Health’s 3D Print Exchange. Researchers at the facility are also using models for teaching, and to help understand the behavior of a large number of different types of molecules.
The facility’s models are designed and output from the PMV molecular modeling environment, and printed on a Z-Corp 510 3D printer.
Because proteins can be very complex, it can be difficult to see how they fold up using 2D renderings. With 3D models, researchers can better examine the molecules and see how individual molecules interact.
For example, Olson’s team has created a fully flexible model of the HIV protease backbone, an important drug target in treating AIDS patients. The model can be folded up from a string of peptide units for each chain. According to the facility’s website: “The folding is accomplished by the interactions of magnets representing hydrogen bond donors and acceptors in the peptide units. The completed model enables manipulation of the two ‘flaps’ that must open to allow the peptide substrate into the active site of the enzyme.”
At the Department of Energy-funded Molecular Foundry at Berkeley Lab in California, nanobioscientist Ron Zuckerman is developing synthetic molecules called peptoids that have the sensitivity of proteins, but can be made of more rugged synthetic amino acids. Researchers there have used 3D printing to better understand how flexible proteins are so they can get a better grasp of how they fold. Forces between molecules can be modeled using magnets, as well as materials with different flexibility to mimic different protein structures
Zuckerman uses printed models of proteins called “peppytides” for educational purposes to show how protein structures form.
Source: Live Science
Subscribe to our FREE magazine, FREE email newsletters or both!
Latest News
About the Author
Brian Albright is the editorial director of Digital Engineering. Contact him at [email protected].
Follow DE